There are a number of research teams working on the problem of skin cancer detection, many of which have near human accuracy within laboratory data sets. There are also a large number of applications released that are used to quantify risk of skin cancer available today. So why am I trying to reinvent the wheel?
Because there is a large gap between the results of academia and the results of industry. The academic papers detailing methods to detect skin cancer are often no more complex than basic object detection or segmentation models. Obviously, companies trying to release these applications for-profit aren’t going to release their models open-source, but I’m assuming they use a similar approach. However, industry results are simply not comparable. SkinVision, one of the leaders in the field of automatic cancer detection, celebrates a 73% detection rate (Study).
My takeaway from these results is that issues start to rise when these models are tested in real world conditions, i.e. when a picture is taken from an iPhone and not a microscope. So my goal with this cancer prediction project is not to build an app that will be more accurate than either the research teams in the lab or than products like SkinVision in industry: I simply don’t have the data or GPU support to make such an endeavor feasible. Instead, my goal is to develop a method of building a data set and training a model that will minimize the difference between accuracy in the lab and accuracy in the real world.
Working with a team of 4 other students at USC, we have a few different routes we areplanning on taking in order to try to gain these results. I am using the publicly available ISIC data set available here: https://isic-archive.com/. This data set contains 13,000 fully labeled images including almost 2000 instances of malignant tumors, and all of these images have been taken under a microscope. (This dataset is extremely difficult to directly download: I ended up using a script I found here.
Route 1: The baseline
With this route, we are going to do a little basic image processioning, and then train a classification layer of a pre-trained model directly on this data, and then evaluate the model against lab data, as well as against a dataset of real world melanoma pictures we scraped together.
Route 2: The Intermediate Path
This is the route I am currently working on myself. I wrote a series of OpenCV scripts that are able to build a mask to isolate the cancer mole within the lab images. Using this mask, I am going to build up a new data set of cancer moles super-imposed on a number of various randomly cropped backgrounds at random lightings, so that the only features our model picks up on are hopefully features within the mole. Hopefully this approach will allow the model to scale better to various backgrounds.

Route 3: Let’s get a little wild 🙂
GANs are cool. Cycle GANs are even cooler. For those of you not familiar with GANs, I really recommend this explanation: An Intuitive Introduction to GANs. Cycle GAN, however, instead of trying to generate something from scratch, is instead trying to transfer 1 image style to another.

However, as you can see, from the picture, this style transfer goes both horses to zebras and zebras to horses: that’s not an accident.
The architecture of the CycleGAN takes the basic GAN architecture of a generator and discriminator trained in parallel- a generator which transfer the style of the image and a discriminator to discern the style.
However, with style transfer, an issue that arises is that the generator, instead of learning a broad method of transfer zebras to horses, may instead learn to convert any picture to the same photo of a single horse. And that would be a problem, because our goal is to keep the new photo as similar to the old as possible while still running a style transfer.
Whenever your results don’t match up with your goal, the first place to look is the loss function. With a regular GAN, our loss function only takes into account whether the style has changed or not. It does not take into account how similar our new photo looks to our original photo.
However, we don’t want to compare our new photo directly to our original photo, since that loss function would result in a high loss even if the style transfer worked perfectly: the new photo will always look different than the original.
So what did smart people do? Well, they threw a GAN on it!
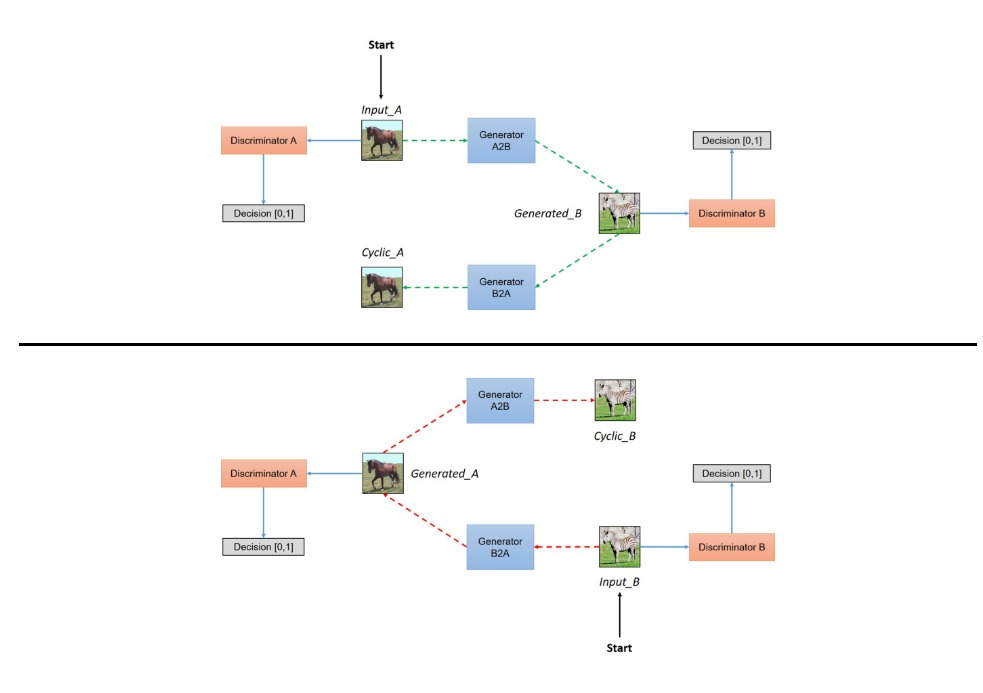
Instead of basing the loss solely on a single discriminator, the model is training another GAN in parallel to convert horses to zebras, whose discriminator is also used to determine the loss.
So the overall flow goes like this: Image of a horse is passed in to original generator, which then outputs the same image post-style-transfer. This image is then passed into the discriminator to determine the loss. However, this image is also passed in as input to another generator: one the converts zebras to horses, which attempts to transfer the previous image back to the original image. The output of this generator is then compared to the original image and the error is then used as the 2nd loss function. As a result, the first generator is trying to generate a zebra in such a way that it fools the discriminator while still retaining enough information to reconstruct the original image.
So, how does this relate to cancer detection?
So I have a theory that, no matter how much you change the background or lighting, the fact that our models are being trained on cancer moles that are themselves taken under the extremely accurate lens of a microscope will seriously impede our model’s ability to accurately detect melanoma in the real-world conditions. So I am hoping to use cycle GAN to run a style transfer on the cancer moles in our lab dataset in order to convert them to real world conditions.
The goal here is to train a model on this on CycleGAN plus background super-imposition data that performs comparably well on real world data as our Baseline model performs on lab data.
Once we have completed these three routes, we will release all our scripts, notebooks, and models on github. The goal is, if this new method works, not only will we be able to release the method, but also we will be able to release a large, self-created dataset of labeled, real-world cancer images derived from the lab images for anyone to use to build whatever model architecture they want. Time to start digging!
